Abstract
The importance of the microbiome in health and its disruption in disease is continuing to be elucidated. However, the multitude of host and environmental factors that influence the microbiome are still largely unknown. Here, we examined UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 3 (Galnt3)-deficient mice, which serve as a model for the disease hyperphosphatemic familial tumoral calcinosis (HFTC). In HFTC, loss of GALNT3 activity in the bone is thought to lead to altered glycosylation of the phosphate-regulating hormone fibroblast growth factor 23 (FGF23), resulting in hyperphosphatemia and subdermal calcified tumors. However, GALNT3 is expressed in other tissues in addition to bone, suggesting that systemic loss could result in other pathologies. Using semiquantitative real-time PCR, we found that Galnt3 is the major O-glycosyltransferase expressed in the secretory cells of salivary glands. Additionally, 16S rRNA gene sequencing revealed that the loss of Galnt3 resulted in changes in the structure, composition, and stability of the oral microbiome. Moreover, we identified the major secreted salivary mucin, Muc10, as an in vivo substrate of Galnt3. Given that mucins and their O-glycans are known to interact with various microbes, our results suggest that loss of Galnt3 decreases glycosylation of Muc10, which alters the composition and stability of the oral microbiome. Considering that oral findings have been documented in HFTC patients, our study suggests that investigating GALNT3-mediated changes in the oral microbiome may be warranted.
Keywords: mucin, glycosylation, microbiome, bone, glycosyltransferase, Galnt3, hyperphosphatemic familial tumoral calcinosis (HFTC), Muc10, O-glycosylation, saliva, salivary protein, glycan, post-translational modification, submandibular gland
Introduction
The importance of the human microbiome throughout the body is only now being fully appreciated. In health, the microbiome interacts synergistically with the host to contribute to normal tissue functions as well as to protect from pathogen colonization (1, 2). Alterations in the composition of the microbiome have been shown to be involved in various disease states, including inflammatory diseases of the digestive tract (3, 4) and diseases of the oral cavity. At this site, dysbiotic changes in the microbiome are associated with dysregulated immune responses that lead to the common oral disease periodontitis (2, 5–7).
The secreted mucosal membrane is a key point of interaction with the microbiome. This protective layer, which is present along most internal epithelial surfaces of the body, forms a barrier that mediates interaction with the microbiota and confers physical protection (8, 9). The main components of this barrier include mucins, proteins with unique rheological properties conferred by the abundant O-glycans present throughout their repetitive domains. O-glycans on mucins bind water to form hydrated gels that line and protect internal epithelial surfaces (8, 9). Additionally, mucins and their associated O-glycans bind microorganisms to modulate clearance, attachment, and the ability to form biofilms (10–12). The importance of mucins and O-glycans in digestive system health is illustrated by studies in model organisms demonstrating that loss of the major intestinal mucin Muc2 or disruption of O-glycan biosynthesis results in spontaneous colitis, colon cancer, and susceptibility to inflammatory conditions of the digestive tract (13–18). However, little is known regarding how individual mucins influence the establishment and composition of the microbiome.
Mucins and O-glycans have been conserved throughout evolution, suggesting important roles in many diverse biological processes (19–22). O-Glycosylation is controlled by a family of enzymes known as the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (Galnts)2 that catalyze the addition of GalNAc to the hydroxyl group of serine or threonine, the first committed step in O-glycan biosynthesis (23, 24). Members of this large family display differential tissue- and cell-specific expression patterns and unique substrate preferences, suggesting that each family member may be responsible for the proper glycosylation of specific proteins in vivo (23, 25, 26). Studies in model organisms have shown that certain Galnts are required for viability and involved in conserved biological processes, such as cell adhesion, secretion, and cell-signaling events (21, 27–34). In humans, mutations in one family member, GALNT3, are responsible for the disease hyperphosphatemic familial tumoral calcinosis (HFTC), characterized by hyperphosphatemia, altered bone density, and the development of subdermal calcified tumors (35). GALNT3-mediated glycosylation of the phosphate-regulating hormone FGF23 within osteocytes of the bone is presumed to protect FGF23 from protease cleavage under normal conditions; loss of the GALNT3 glycosyltransferase results in inactivating cleavage of FGF23 and dysregulation of phosphate homeostasis. A mouse model of this disease that is deficient for Galnt3 recapitulates many disease phenotypes, including Fgf23 inactivating cleavage and hyperphosphatemia (36). Whereas both males and females displayed these HFTC hallmark phenotypes, only males displayed growth retardation, infertility, and increased bone density (36, 37). The reasons for these gender-specific differences are currently unknown.
Interestingly, GALNT3 (Galnt3 in mice) is abundantly expressed in other tissues and thus may have a role in other parts of the body that could contribute to disease pathology (21, 23). Here, we identify Galnt3 as the major family member expressed in adult mouse salivary glands, which are responsible for producing the components of saliva, including mucins. Interestingly, we show that loss of Galnt3 results in dramatic changes in the composition and structure of the oral microbiome. We further identify the major secreted mucin Muc10 as an in vivo and in vitro substrate of Galnt3. Our results demonstrate for the first time that the loss of a single glycosyltransferase can alter the oral microbiome, possibly through altered glycosylation of a major salivary mucin. Given the importance of the microbiome in health and disease, our study suggests that investigation of the oral microbiome in patients with mutations in GALNT3 may be warranted to have a more comprehensive understanding of disease pathology.
Results
Galnt3 is the most abundant Galnt in the adult submandibular gland
Previous studies have shown that Galnt3 is abundantly expressed in other tissues throughout the body, including the salivary glands (23, 38). To begin to interrogate the relative abundance and temporal expression of Galnt3 in the submandibular glands (SMGs), we investigated expression levels of each Galnt family member at three embryonic stages (embryonic day 13 (E13), E15, and E17), two early postnatal stages (postnatal day 2 (P2) and 1 week), and three adult stages (4, 8, and 12 weeks) by performing qPCR (Fig. 1 and Fig. S1). At E13, Galnt1 is the most abundantly expressed isoform, followed by Galnt2 and Galnt11 (Fig. 1A). At E15 and E17, Galnt2 becomes the most abundantly expressed isoform, followed by Galnt1 at E15 and Galnt4 and Galnt1 at E17 (Fig. 1A). Interestingly, a dramatic change in Galnt expression occurs after birth, as Galnt3 and Galnt12 are the most abundantly expressed isoforms at P2 and 1 week of age (Fig. 1B and Fig. S1A). Moreover, Galnt3 expression continues to increase into adulthood, where it is by far the most abundant isoform at 4, 8, and 12 weeks of age (Fig. 1B and Fig. S1A), in both males and females (Fig. S1B). In situ hybridization revealed that Galnt3 expression is confined to the acinar cells (detected with Aqp5) of the SMGs, the cells responsible for producing the secreted components of the saliva (Fig. 1C).
Figure 1.
Galnt3 is the most abundantly expressed isoform in adult SMGs. A, expression of Galnt family members in embryonic SMGs detected by qPCR. Galnt1 is the predominant isoform at E13, and Galnt2 is the predominant isoform at E15 and E17. B, expression of Galnt family members (with greater than 2% relative expression) in early postnatal and adult SMGs. Expression of entire murine Galnt family is shown in Fig. S1A. Galnt3 is the most abundant isoform in adult SMGs. Values represent mean ± S.D. (error bars) from three or more animals. Expression was normalized to 29S rRNA and is represented as percentage of total Galnt expression for each individual stage of development. C, in situ hybridization using TSA in combination with immunofluorescent staining of 8-week SMGs shows that Galnt3 mRNA (red) is found specifically in acinar cells (detected by Aqp5, cyan) in both males and females. Nuclear staining is shown in blue. Scale bar, 10 μm.
Previous work has shown that one of the most abundantly expressed Galnts during embryonic stages (Galnt1) plays key roles during SMG growth and development (28). This led us to examine the possibility that Galnt3, the most abundant isoform in adult SMGs, may play a role in adult SMG function. To investigate this, we crossed heterozygous Galnt3-deficient (Galnt3+/−) animals to generate homozygous Galnt3-deficient (Galnt3−/−) animals and WT littermate controls. Galnt3−/− mice have been previously generated as a mouse model for HFTC and were found to have hyperphosphatemia, inappropriately normal levels of 1,25-dihydroxyvitamin D, and decreased levels of circulating intact Fgf23, similar to what is seen in HFTC patients (36). As it is well-documented that rodent SMGs become morphologically distinct between males and females beginning at ∼6 weeks of age (with males developing many more granular convoluted tubules than females) (39), all future experiments analyzed male and female animals separately. Upon examination of SMGs at 8 weeks of age, we found no significant differences in gland weight (Fig. S2, A and B) or morphology (Fig. S2C) between WT and Galnt3−/− mice for either sex. Previous studies from our laboratory investigating the role of another member of this family in the SMG found an induction of ER stress upon loss of Galnt1 (28). To determine whether the loss of Galnt3 also induced ER stress and the unfolded protein response (similar to that seen in Galnt1−/− SMGs (28)), we next examined Xbp1 mRNA splicing. Quantification of the ratio of spliced Xbp1 (Xbp1s) to unspliced Xbp1 (Xbp1u) revealed no statistically significant differences between WT and Galnt3−/− SMGs for either sex (Fig. S2, D–G), indicating that the loss of Galnt3 does not result in ER stress. Additionally, we performed a number of analyses of adult SMG function. Ex vivo analysis of SMG salivary flow rate, volume of saliva secreted, and ion content of saliva revealed no significant differences between WT and Galnt3−/− animals for either sex (Fig. S3, A–F). These results indicate that the loss of Galnt3 does not result in overt differences in SMG size, morphology, or function via these assays.
Loss of Galnt3 alters the oral microbiome
In addition to its role in hydration and lubrication of the oral cavity, saliva is believed to play a role in regulating the oral microbiota (40). This led us to investigate the effect of loss of Galnt3 on the microbial communities within the oral cavity through the characterization of the local microbiome. Microbiome studies analyze the numbers and types of distinct microbial taxa as well as their relative abundances to provide assessments of microbial community structures. We collected oral mucosal samples as described previously (41) from individual WT and Galnt3−/− mice at both 8 and 12 weeks of age and performed 16S rRNA gene sequencing. β-diversity comparisons of the microbial community structure within the oral cavity across genotypes and age groups were assessed using the θ Yue and Clayton (θ YC) distance (which takes into account the specific bacterial species present as well as their relative abundances within the communities being compared) and visualized on two-dimensional principal coordinates analysis (PCoA) plots (Fig. 2). We also analyzed α-diversity within each genotype and age group (which measures the number of bacterial taxa and their relative proportions within a particular group), using the nonparametric version of the Shannon index (Fig. S4).
Figure 2.
Loss of Galnt3 alters the oral microbiome. Shown are PCoA plots of oral swab samples based on θ YC distances (to analyze microbial community structure) with 95% confidence ellipses. The oral microbiome was sampled at both 8 and 12 weeks for each individual animal. For both 8-week-old (A) and 12-week-old (B) males, WT samples (red dots) cluster apart from Galnt3−/− samples (gray dots). Each dot represents an individual mouse. At both 8 and 12 weeks of age, statistically significant differences between WT and Galnt3−/− samples were determined by AMOVA (p < 0.05). For females, no significant differences between WT (brown dots) and Galnt3−/− (pink dots) samples were observed at 8 weeks (C) or 12 weeks of age (D). Shown is the relative abundance of bacterial taxa at genus level in oral swab samples from 8-week-old (E) and 12-week-old (F) males. Each bar represents one mouse. Unclassified genera are shown at the family level. Species level taxonomy is reported in parenthesis when >97% similarity was achieved using NCBI BLAST. *, species overrepresented in WT samples; #, species overrepresented in Galnt3−/− samples according to LEfSe analyses.
Through β-diversity analysis, we found distinct microbial communities in Galnt3−/− male samples compared with WT at both 8 and 12 weeks of age (Fig. 2, A and B). PCoA plots demonstrate that there was significant clustering and separation of male Galnt3−/− microbial communities from male WT communities (p < 0.05 as determined by analysis of molecular variance (AMOVA); Fig. 2, A and B). In contrast to male samples, no significant differences between the community structure of WT and Galnt3−/− female samples were observed for either age (Fig. 2 (C and D) and Fig. S5 (A and B)). No significant differences in α-diversity were observed for either males or females (Fig. S4, A–D). These results indicate that there are significant microbial shifts in the oral microbiome community structure in males upon loss of Galnt3.
Examining the composition of the oral microbiome in more detail, we found specific changes in the relative abundance of select taxa between male WT and Galnt3−/− animals. At 8 weeks of age, two operational taxonomic units (OTUs) from the Pasteurellaceae family (which had a high percentage of similarity with the species Muribacter muris and Rodentibacter heidelbergensis), were overrepresented in WT samples, whereas a Lactobacillus sp. (Lactobacillus faecis) and two other low-abundance OTUs (an unclassified Lachnospiraceae sp. and a Candidatus Saccharibacteria/TM7 sp.) were increased in Galnt3−/− samples (Fig. 2E). At 12 weeks of age, a Streptococcus sp. (Streptococcus danieliae) was overrepresented in WT samples, whereas a Pasteurellaceae sp. (R. heidelbergensis) became overrepresented in Galnt3−/− samples (Fig. 2F). Interestingly, 12-week Galnt3−/− samples continued to show an increase in Lactobacillus sp. (L. faecis) as compared with WT (Fig. 2F). It is notable that these data come from mice of both genotypes housed across multiple cages, eliminating the possibility that the distinct microbial communities are due to cage-specific effects. We next examined the stability of the oral microbiome in individual adults of each genotype over time. Similar to previous studies in mice and humans (6, 42), the microbial community structure of WT animals was stable over time, as no significant differences were found between 8- and 12-week samples for either male or female animals (Fig. 3 (A and B) and Fig. S5 (C and D)). Likewise, no significant differences were seen in α-diversity (Fig. S4, E and F). However, significant differences in community structure were seen over time in Galnt3−/− animals (Fig. 3, C and D). For both males and females, 8-week Galnt3−/− oral mucosal communities significantly separated from 12-week Galnt3−/− communities, and these differences were found to be statistically significant by AMOVA (p < 0.05) (Fig. 3, C and D). Investigating age-related differences in the relative abundance of bacterial taxa in more detail in male Galnt3−/− samples, we found that the OTUs Pasteurellaceae sp. (M. muris) and Pasteurellaceae sp. (R. heidelbergensis) were overrepresented in 12-week samples as compared with 8 weeks (Fig. 3E). Similarly, these two OTUs belonging to the Pasteurellaceae family were also more abundant in 12-week female Galnt3−/− samples as compared with 8 weeks, whereas a Streptococcus sp. (S. danieliae) was overrepresented at 8 weeks (Fig. 3F). Whereas there were no significant changes in α-diversity in Galnt3−/− males over time, there was a significant increase in α-diversity in 12-week Galnt3−/− females (Fig. S4, G and H). These results indicate that the composition of the oral microbiome is less stable over time upon loss of Galnt3 in both males and females.
Figure 3.
Loss of Galnt3 alters stability of the oral microbiome over time. Shown are PCoA plots comparing 8- and 12-week oral swab samples based on θ YC distances (to analyze microbial community structure) with 95% confidence ellipses. The oral microbiome was sampled at both 8 and 12 weeks for each individual animal. For both WT males (A) and females (B), no significant differences between 8- and 12-week samples were observed. For Galnt3−/− animals, 8-week samples cluster apart from 12-week samples for both males (C) and females (D). Statistically significant differences between 8- and 12-week Galnt3−/− samples were determined by AMOVA (p < 0.05). Shown is the relative abundance of bacterial taxa at the genus level in oral swab samples from male (E) and female (F) Galnt3−/− animals. Each bar represents one mouse. Unclassified genera are shown at the family level. Species-level taxonomy is reported in parenthesis when >97% similarity was achieved using NCBI BLAST. *, species overrepresented in 12-week samples; #, species overrepresented in 8-week samples according to LEfSe analyses.
We also compared the microbiomes between WT males and females at each time point. Whereas a difference in structure was seen at 8 weeks of age (Fig. S6A), this difference was no longer significant by 12 weeks of age (Fig. S6B). Examination of the taxonomy data reveals that the differences at 8 weeks of age are driven by differences in the relative abundance of the major taxa rather than the presence or absence of unique taxa (Fig. S6, C and D). Likewise, there were no significant differences in α-diversity between WT males and females at either age (Fig. S4, I and J).
Galnt3 glycosylates Muc10
To investigate how the loss of Galnt3 (which encodes an O-glycosyltransferase that catalyzes the first committed step in O-glycan biosynthesis) may be influencing the composition and stability of the microbiome, we next examined the O-glycans that are normally present within the SMG using lectins that are specific to O-glycan structures. Previous studies have documented distinct gender differences in the expression of the ST3Gal1 gene (which encodes a sialyltransferase that adds sialic acid in an α2,3 linkage to glycans), with this gene being 22-fold more abundantly expressed in the SMGs of females versus males ((https://sgmap.nidcr.nih.gov/cgi-bin/sgexp11.plx?ind=1&gn=St3gal1). Indeed, female SMGs stained abundantly with the lectin Maackia amurensis agglutinin (MAA) (Fig. 4A), which detects α2,3-linked sialic acid linked to either N- or O-glycans (43). This staining was seen in the acinar cells of the SMG (labeled with the marker acinar-1) and was specifically removed upon treatment with an enzyme that removes sialic acid (neuraminidase (NM)), indicating specificity (Fig. 4A). Staining with MAA was also seen in male SMG acinar cells, but to a lesser extent (Fig. 4B). We next used the lectin peanut agglutinin (PNA) (which detects the nonsialylated core 1 structure Galβ1,3GalNAc of O-glycans) in the presence or absence of NM treatment to determine the degree to which O-glycans are sialylated in both WT females and males (Fig. 4, C and D). As shown in Fig. 4D, PNA staining is abundantly present within the acinar cells (labeled with the acinar-1 marker) of male SMGs with or without NM treatment, indicating the presence of nonsialylated core 1 O-glycans in males. However, PNA staining was only detected in female SMGs after NM treatment, indicating that all core 1 O-glycans are modified with sialic acid in females (Fig. 4C). PNA staining in both males and females could be competed away by incubation with galactose, indicating specificity (Fig. S7). Similar results were obtained on western blots of SMG extracts, where female samples showed no PNA reactivity in the absence of NM treatment, whereas male samples had PNA reactivity with or without NM treatment (Fig. 4, E and F). Taken together, these results indicate that distinct differences exist between males and females in the structure of the O-glycans present within the SMGs, with males having both sialylated and nonsialylated core 1 O-glycans and females having core 1 O-glycans that are predominantly sialylated. Therefore, all further analysis of PNA-reactive O-glycans in females was performed on samples that had been treated with NM, whereas male samples were not treated with NM.
Figure 4.
Glycans present within the acinar cells of SMGs. Immunofluorescent staining of 8-week WT female (A) and male (B) SMGs with the lectin MAA (green), which detects α2,3-linked sialic acid on O- and N-linked glycans. Samples were either untreated (No NM) or treated with neuraminidase (NM), which removes sialic acid to show specificity. Acinar cells are shown with the marker acinar-1 (red). Nuclei are shown in blue. Scale bars, 20 μm. Shown is immunofluorescent staining of 8-week WT female (C) and male (D) SMGs with the lectin PNA (green), which detects the nonsialylated core 1 O-glycan Galβ1,3GalNAc. Samples were either untreated (No NM) or treated with neuraminidase (NM). No PNA staining is seen in untreated female SMGs, whereas PNA staining is seen in both treated and untreated male SMGs. Acinar cells are shown with the marker acinar-1 (red). Nuclei are shown in blue. Scale bars, 10 μm. Shown are western blots of WT female (E) and male (F) SMG extracts either untreated (−) or treated with NM (+) and probed with PNA. Molecular mass markers (kDa) are shown to the left of each blot.
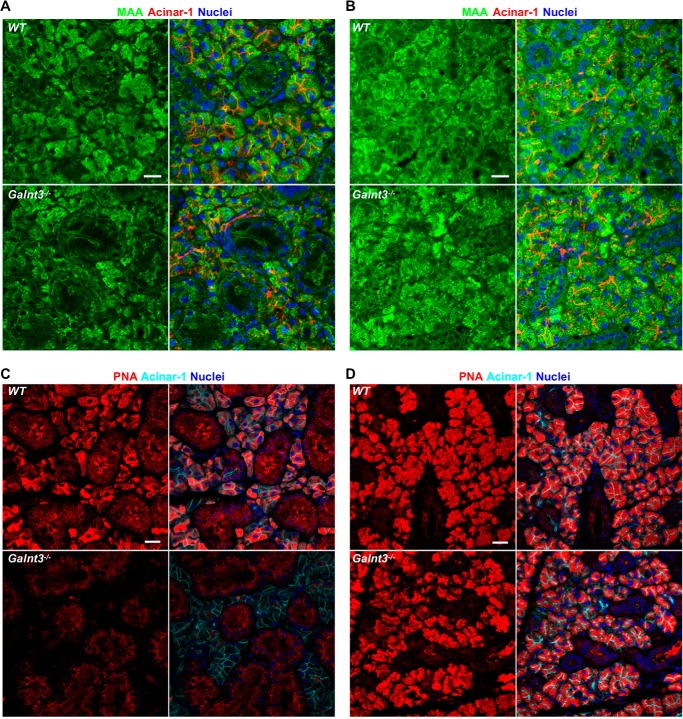
We next set out to compare changes in glycans present in WT and Galnt3−/− SMGs via confocal microscopy. As shown in Fig. 5 (A and B), loss of Galnt3 did not result in obvious changes in MAA staining in either males or females. However, a dramatic decrease in PNA-reactive O-glycans specifically in the acinar cells of male SMGs was seen upon loss of Galnt3 (Fig. 5C). In contrast to males, female Galnt3−/− SMGs (treated with NM) did not show a noticeable change in PNA-reactive O-glycan staining within the acinar cells relative to WT via confocal imaging (Fig. 5D). These results suggest that the loss of Galnt3 results in a major change in PNA-reactive O-glycans normally present within the acinar cells of male SMGs.
Figure 5.
Loss of Galnt3 results in diminished O-glycans in male acinar cells. Immunofluorescent staining of 8-week WT and Galnt3−/− male (A) and female (B) SMGs with the lectin MAA (green), which detects α2,3-linked sialic acid on O- and N-linked glycans. Acinar cells are shown with the marker acinar-1 (red). Nuclei are shown in blue. Scale bars, 10 μm. C, immunofluorescent staining of 8-week male SMGs shows a dramatic reduction in O-glycans (detected by PNA, red) specifically in the acinar cells (detected by acinar-1, cyan) of Galnt3−/− SMGs as compared with WT. D, immunofluorescent staining of neuraminidase-treated 8-week female SMGs reveals no obvious differences in PNA-reactive O-glycans between WT and Galnt3−/− SMGs. Nuclear staining is shown in blue. Scale bars, 20 μm.
We next set out to identify the potential protein targets of Galnt3 in the SMGs. Mucins are heavily O-glycosylated proteins known to be the major protein products produced by the salivary glands and are thought to mediate interactions with the microbiome (12, 44). Previous studies have identified mucin 10 (Muc10; also known as mouse submandibular gland mucin and proline-rich, lacrimal 1) as the major mouse SMG mucin and have shown that it localizes to the acinar cells of adult SMGs (45, 46). Here, we examined in detail the postnatal gene expression of Muc10 using qPCR (Fig. 6A). At all five stages examined (P2, 1 week, 4 weeks, 8 weeks, and 12 weeks), Muc10 was found to be abundantly expressed, with its expression increasing in adult stages (Fig. 6A). qPCR to other mucins revealed that Muc10 was by far the most abundantly expressed mucin within the SMGs (Fig. 6, A–D). Additionally, we confirmed that Muc10 is also expressed in acinar cells (detected with acinar-1) of SMGs (Fig. 6E) similar to Galnt3, suggesting that Muc10 may be a direct target of Galnt3.
Figure 6.
Analysis of mucin gene expression in early postnatal and adult SMGs. A, qPCR analysis demonstrates that Muc10 is the most abundantly expressed mucin gene at all five stages, with increasing expression in adult stages. Moderate to low levels of expression were detected for Muc13 (B), Muc20 (C), and Muc1 (D). Muc2, Muc4, Muc5ac, and Muc6 were not detected at any stage. Values represent mean ± S.D. (error bars) from three or more animals. Expression was normalized to 29S rRNA. E, immunofluorescent staining of 8-week SMGs shows specific localization of Muc10 protein (red) to acinar cells (detected by acinar-1, cyan) in both males and females. Nuclear staining is shown in blue. Scale bar, 20 μm.
Given the abundant expression of Muc10 within acinar cells of the SMGs, we next set out to determine whether O-glycosylation of Muc10 was specifically affected by the loss of Galnt3. We performed western blot analysis of 8-week WT and Galnt3−/− SMG extracts, probing for PNA and Muc10 (Fig. 7). In WT male SMGs, two major PNA bands were detected (Fig. 7A). However, in Galnt3−/− SMGs, the lower-molecular weight PNA-reactive band was greatly reduced or absent, suggesting that this protein is specifically glycosylated by Galnt3. Interestingly, probing the same western blot for Muc10 revealed that the Muc10 band overlaps with the lower PNA-reactive band in WT SMGs, suggesting that Muc10 may be this highly O-glycosylated band (Fig. 7A). In support of this, immunoprecipitation of Muc10 from WT SMGs revealed that it does contain PNA-reactive O-glycans (Fig. S8). Additionally, the Muc10 band in Galnt3−/− SMGs no longer overlaps with PNA staining and runs at a reduced size, further suggesting the loss of O-glycans on this protein (as these sugar modifications can account for a significant change in molecular weight). These results strongly suggest that Muc10 is specifically O-glycosylated by Galnt3 in male SMGs. western blots of 8-week female SMGs did not reveal the same PNA banding pattern seen in WT males and did not result in the loss of specific PNA-reactive bands, as seen in male Galnt3−/− SMGs (Fig. 7B). However, the loss of Galnt3 in female SMGs did result in PNA reactivity with altered mobility (Fig. 7B). Additionally, Muc10-reactive bands ran as large smears that overlapped in position with PNA staining in Galnt3−/− female SMGs (Fig. 7B). These results suggest that Muc10 is also a substrate for Galnt3 in female SMGs and that, while the loss of Galnt3 does not result in a complete disruption of O-glycosylation (as detected by PNA), it may alter the patterns of O-glycosylation in more subtle ways in females.
Figure 7.
Muc10 is an in vivo substrate for Galnt3. A, western blots of 8-week male SMG lysates probed for O-glycans (detected by PNA, red) and Muc10 (green). Separated channels are shown in black in the top two panels, and merged channels are shown in color in the bottom panel. The lower of the two PNA-reactive bands overlaps with the Muc10 band in WT SMGs. In Galnt3−/− SMGs, the lower PNA-reactive band is absent, and the Muc10 band is smeared and reduced in size. B, western blots of neuraminidase-treated 8-week female SMG lysates probed for O-glycans (detected by PNA, red) and Muc10 (green). The PNA and Muc10 bands overlap in both WT and Galnt3−/− SMGs. In Galnt3−/− SMGs, both the PNA and Muc10 bands exhibit altered mobility as compared with WT. Molecular mass markers (kDa) are shown to the left of each blot. C, in vitro enzymatic activity of Galnt3 against Muc10 acceptor peptides (peptide sequences shown in D). EA2 was used as a positive control. Galnt3 was able to glycosylate the Muc10-s, Muc10-273, Muc10-264, and Muc10-l peptides. Galnt3 showed no activity toward the Muc10-10 peptide. Each data point represents an individual assay. Error bars, S.D.
To further investigate the ability of Galnt3 to glycosylate Muc10, we performed in vitro enzymatic assays using recombinant murine Galnt3 and acceptor peptides from Muc10. Standard transferase assays revealed that Galnt3 was able to glycosylate four different peptides derived from Muc10 (Fig. 7, C and D), providing further evidence that Muc10 is a substrate for Galnt3 in the SMGs. Interestingly, Galnt3 had specific site preferences among the Muc10 peptides, showing highest activity toward the Muc10-s and Muc10-264 peptides, followed by Muc10-273 and Muc10-l (all of which are derived from the repetitive domain of Muc10). Galnt3 did not show activity toward the Muc10-10 peptide (Fig. 7 (C and D) and Figs. S9 and S10). These results are in agreement with predicted site preferences previously determined for Galnt3 (47). Taken together, our results demonstrate that loss of Galnt3 leads to changes in the composition of the oral microbiome in males, possibly due to changes in the glycosylation status of the major salivary mucin Muc10. Moreover, loss of Galnt3 results in altered stability of the oral microbiome in both males and females. Given that mutations in GALNT3 cause the human disease HFTC and that Galnt3−/− mice serve as a model for this disease, our results suggest that HFTC patients may also have alterations in their oral microbiome that could contribute to documented disease pathology in the oral cavity (Table S1).
Discussion
Here, we demonstrate that the gene responsible for HFTC alters the composition and stability of the oral microbiome. Galnt3 is the most abundantly expressed member of the Galnt family in the acinar cells of adult SMGs, which produce the components of the saliva that lines and protects the oral cavity. Loss of Galnt3 resulted in dramatic changes in the predominant bacterial taxa present, as well as significant changes in the overall composition of the oral microbiome in males. Moreover, we found that while the composition of the oral microbiome was stable over time in WT, this stability was lost in both male and female Galnt3-deficient animals, suggesting that the presence of Galnt3-initiated glycans confers stability to the endogenous microbial communities. Previous studies have documented the notable stability of the oral microbiome in association with health (42), as well as how dysbiotic changes in the oral microbiome can trigger an increase in inflammation leading to periodontitis (2, 5–7). Indeed, bacterial homoeostasis is a crucial part of oral health, as several prevalent oral diseases such as dental caries, periodontitis, and oral candidiasis are associated with local microbial dysbiosis (48, 49). Additionally, changes in the oral microbiome in certain genetic disorders underlie local inflammation and pathology observed within the oral cavity (50, 51). As some clinical reports of HFTC patients have included oral findings (Table S1), our results in the mouse model of this disease may provide insight into how the systemic loss of GALNT3 may result in pathology within the oral cavity.
We further demonstrate that changes in the composition of the oral microbiome upon loss of Galnt3 are associated with changes in the glycosylation status of the major salivary mucin, Muc10. Whereas there were no overt phenotypic or functional differences between WT and Galnt3−/− SMGs, there was a significant loss of O-glycans within the acinar cells of male Galnt3−/− glands. This change in glycosylation was in part due to dramatic changes in the glycosylation status of Muc10 in males. Moreover, we show that Galnt3 glycosylates specific regions of Muc10 in vitro, in accordance with predicted site preferences based on bioinformatic analyses, establishing Muc10 as an in vivo and in vitro substrate of Galnt3. Interestingly, we did not see significant changes in the abundance of O-glycans in female Galnt3−/− SMGs, just as we did not see significant changes in the composition of the oral microbiome in females. However, the females did display changes in Muc10 mobility, suggesting more subtle changes in its glycosylation pattern. It is worth noting that the core 1 O-glycans present in WT SMGs differ between males and females, with female O-glycans being predominantly capped with sialic acid. How gender-specific differences in the degree of sialylation of proteins may influence various biological processes remains to be investigated.
Our study supports a model where altered glycosylation of a major salivary mucin may lead to alteration and instability of the oral microbiome, which has physiological implications regarding host-microbial interactions at this interface. This model is further supported by many studies documenting the interaction of bacteria with mucins and O-glycans, influencing their clearance and ability to colonize (10, 11). Additionally, glycans on mucins can interact with bacterial receptors to influence microbial behavior and signaling (52, 53). Interestingly, the human ortholog of Muc10 (MUC7/MG2) has documented roles in bacterial binding and aggregation (54–56), suggesting that this mucin may play a role in maintaining the stability of the oral microbiome in health. Indeed, a recent study demonstrated that a human oral microbe recognizes α2,3-sialylated core 1 structures on the human ortholog of Muc10 (MUC7) (57), highlighting the importance of sialylated O-glycans in bacterial recognition. Given the dramatic differences in the degree of sialylation of O-glycans seen between males and females, it will be interesting to investigate gender-specific glycosylation patterns and how they may influence biological phenotypes.
In summary, we have found changes in the composition and stability of the oral microbiome in the mouse model of the human disease HFTC. Whether HFTC patients (who lack functional GALNT3) also have changes in the composition of their oral microbiome is currently unknown. However, GALNT3 is abundantly expressed in human SMGs (21), and a number of clinical studies have noted oral findings in HFTC patients, including caries, gingivitis, periodontitis, and changes in enamel (Table S1). Given our results and the obvious importance of the microbiome in health and organ function, our study suggests that examining the oral microbiome and oral phenotypes in HFTC patients may be informative.
Experimental procedures
Animal breeding and genotyping
The Galnt3-deficient mice were a kind gift from Dr. Michael J. Econs (36). Galnt3-deficient mice were backcrossed into the C57BL/6MHsd inbred mouse background for at least six generations before analysis. Heterozygous Galnt3 animals (Galnt3+/−) were crossed to generate WT and homozygous Galnt3-deficient (Galnt3−/−) siblings to be used for all experiments. Genomic DNA was extracted from mouse ear punches using the DNeasy Blood & Tissue Kit (Qiagen #69506), and PCR was performed as described previously (36). Experimental procedures were reviewed and approved by the Animal Care and Use Committee of the National Institutes of Health (ASP #17-833).
Semiquantitative real-time PCR
RNA was isolated and purified using the PureLinkTM RNA Mini Kit (Invitrogen #12183018A). cDNA was synthesized using iScriptTM Reverse Transcription Supermix (Bio-Rad). Primers for Galnt and Mucin genes were designed using Beacon Designer software and are shown in Tables S2 and S3, respectively. qPCR was performed for 40 cycles (95 °C for 10 s and 62 °C for 30 s) using SYBR Green PCR Master Mix, either 1 ng (Galnt genes) or 10 ng (Mucin genes) of each cDNA sample, and the CFX96 real-time system (Bio-Rad). Gene expression was normalized to 29S rRNA. Reactions were run in triplicate and repeated using three or more animals.
Histology
8-Week mouse submandibular glands were fixed in 4% PFA/PBS overnight at 4 °C, transferred to 70% ethanol, and stored at 4 °C. Paraffin-embedded tissue sections (5 μm) were used for hematoxylin/eosin staining, in situ hybridization, and immunofluorescent staining.
In situ hybridization
Primers of Galnt3 probes were designed using Beacon Designer software. Probes were amplified by the KOD kit (TAKARA) and purified by a QIAquick spin kit (Qiagen). Digoxigenin-11-UTP–labeled single-stranded RNA probes were prepared using a DIG RNA labeling kit (Sigma-Aldrich #11175025910). Fixed, paraffin-embedded SMG sections (5 μm) were dewaxed at 60 °C for 10 min followed by three changes of xylene substitute (Sigma #A5597). Sections were rehydrated with two changes of 100% ethanol and single changes of 90, 70, and 50% ethanol. Sections were fixed by 4% PFA/PBS for 5 min, followed by 10 mg/ml Proteinase K (Sigma #4850) treatment for 30 min at 37 °C. After post-fixing with 4% PFA/PBS for 5 min, sections were dehydrated in ethanol series (50, 70, 90, and 100%). Hybridization was performed by incubating sections with 50 ng of Galnt3 RNA DIG-labeled antisense or sense probes in 25 μl of hybridization buffer for 10 min at 85 °C and then overnight at 50 °C. After washing the next day, sections were treated with 10 mg/ml RNase A in 10 mm Tris-HCl (pH 7.5), 500 mm NaCl for 15 min at 37 °C. Following washing, sections were blocked with 1% blocking reagent (Roche Applied Science #1096176) for 30 min at room temperature and then incubated with anti-DIG-POD antibody (Roche Applied Science; 1:100) and Aquaporin-5 primary antibody (Calbiochem #178615; 1:100) overnight at 4 °C. On day 3, sections were washed and then incubated with Alexa Fluor 647®-conjugated anti-rabbit IgG (Jackson ImmunoResearch; 1:400) for 1 h at room temperature. Following washing, sections were incubated with tyramide signal amplification plus Cy3 (TSA; PerkinElmer Life Sciences; 1:50 in amplification buffer) for 30 min at room temperature. After washing, sections were nuclear-counterstained with Hoechst 33342 (Invitrogen #H3570; 1:10,000) and mounted in aqueous Fluro-Gel medium (Electron Microscopy Sciences #17985-10). Sections were visualized on a Nikon A1R confocal microscope, and images were processed in Fiji (National Institutes of Health).
Immunofluorescent staining
Fixed, paraffin-embedded tissue sections (5 μm) were deparaffinized by incubating slides at 60 °C for 10 min, followed by three washes in xylene substitute (Sigma #A5597). Sections were rehydrated with single changes of 100, 90, 70, 50, and 30% ethanol and deionized water. Antigen retrieval was performed by incubating slides in 5% urea and 50 mm β-mercaptoethanol for 10 min at 95 °C, followed by 40 min of cooling. For female samples to be probed with PNA (except where noted), sections were treated with 20 milliunits of neuraminidase (New England BioLabs #P0720) overnight at 37 °C. Sections were blocked with 10% donkey serum, 1% BSA, and M.O.M blocking reagent (Vector Laboratories #FMK-2201). Where noted, some male samples were also treated with neuraminidase via the same procedure. Primary antibodies were diluted in PBS, 0.1% Tween 20 plus M.O.M. protein reagent and incubated overnight at 4 °C for male samples or for 2 h at room temperature for female samples. Primary antibodies used were Muc10 (Everest Biotech #EB10617; 1:200) and acinar-1 (Developmental Studies Hybridoma Bank #3.7A12; 1:20). After washing, sections were incubated for 1.5 h at room temperature with either TRITC-conjugated MAA (EY Laboratories #R-7801-2; 1:100) or FITC-conjugated PNA (Vector Laboratories #FL-1071; 1:200), CyTM3-conjugated anti-goat IgG (Jackson ImmunoResearch; 1:400), and Alexa Fluor 647®-conjugated anti-rat IgG (Jackson ImmunoResearch; 1:400). For sugar inhibition of PNA, 0.2 m galactose (Sigma-Aldrich #G0750) was incubated with FITC-labeled PNA for 1 h at room temperature before applying to slides. Nuclear counterstaining was done using Hoechst 33342 (Invitrogen #H3570; 1:10000) for 3 min at room temperature. Stained sections were mounted in aqueous Fluro-Gel medium (Electron Microscopy Sciences #17985-10) and visualized on a Nikon A1R confocal microscope. Images were processed in Fiji, and representative stacked section confocal images are shown in each figure. FITC-labeled PNA was pseudocolored to red, and TRITC-labeled MAA was pseudocolored to green in Fiji (Figs. 4 (A and B) and 5 and Fig. S6).
Immunoprecipitation
Immunoprecipitation was performed using TrueBlot® anti-goat IgG magnetic beads (Rockland Immunochemicals Inc. #00-1844) according to the manufacturer's instructions with modifications. 8-Week WT SMG lysates were prepared in RIPA buffer with sonication. 25 μg of SMG lysates were combined with 5 μg of Muc10 antibody (Everest Biotech #EB10617), diluted to 250 μl in RIPA buffer, and incubated overnight at 4 °C with mixing. The next day, beads were washed with RIPA buffer. 200 μl (1 mg) of prewashed beads were then combined with the preformed SMG lysate/antibody complexes and incubated overnight at 4 °C with mixing. Next, beads were collected, and the supernatant was removed, followed by washing with RIPA buffer. Beads were then resuspended in RIPA buffer and SDS-loading dye (containing a 1:25 dilution of β-mercaptoethanol) and heated at 95 °C for 10 min, followed by cooling on ice for 10 min. The supernatant (containing the target antigen) was then removed from the beads and used for SDS-PAGE and western blotting as described below.
Western blotting
PNA (Arachis hypogaea lectin, EY Laboratories #L-2301) was labeled with IRDye 680LT (LI-COR Biosciences #928-38066). 8-Week SMGs from littermate WT and Galnt3−/− animals were lysed in RIPA buffer with sonication. SMGs from male and female animals were treated with 6.25 milliunits of neuraminidase where noted (New England BioLabs #P0720) for 1 h at 37 °C. Samples were analyzed by SDS-PAGE under denaturing and reducing conditions and transferred to nitrocellulose membranes. Membranes were blocked with 1:1 PBS/Odyssey Blocking Buffer (LI-COR Biosciences #927-4000) and probed with Muc10 (Everest Biotech #EB10617; 1:2000). After washing, membranes were probed with IRDye 680LT-conjugated PNA (1:10000) and IRDye 800CW-conjugated anti-goat IgG (LI-COR Biosciences #925-32214; 1:5000). Membranes were scanned using a LI-COR Odyssey IR imaging system.
Xbp1 mRNA splicing PCR
PCR primers used to detect splicing of Xbp1 were published previously (28). PCR was performed for 35 cycles (94 °C for 30 s, 61 °C for 30 s, 72 °C for 90 s) using 10 ng of each cDNA sample and a VeritiTM thermal cycler (Applied Biosystems). PCR products were identified using 4–20% TBE gels and ethidium bromide. Negative images of gels are shown. NIH Fiji was used to measure band intensity, and normalized ratios of spliced Xbp1/unspliced Xbp1 were determined (ratio for WT controls set to 1).
Enzyme assays
Expression of recombinant murine Galnt3 was performed as described previously (38) using COS7 cells. Assays for Galnt3 activity were performed as described previously (38) using 5 μl of media from COS7 cells transfected with murine Galnt3. Muc10 and EA2 peptide substrates were synthesized by Peptide 2.0 (Fig. 5D and Fig. S6). Reactions were run for 1.5 h at 37 °C and then quenched with 30 mm EDTA. Glycosylated products were separated from unincorporated UDP-[14C]GalNAc by anion-exchange chromatography using AG1-X8 resin columns (Bio-Rad #1401454), and product incorporation was determined by liquid scintillation counting. Background values were obtained from reactions using media from COS7 cells transfected with empty vector and subtracted from each experimental value. Assays for each peptide substrate were run in triplicate and repeated three times.
Ex vivo perfused SMG
Ex vivo SMG saliva secretions were collected by modifying the ex vivo mouse SMG perfusion technique (58). In brief, mice were anesthetized by chloral hydrate injection (400 mg/kg intraperitoneally), and all branches of the common carotid artery were ligated, except for the artery supplying blood to SMG. Intact glands were then removed and transferred to a temperature-controlled perfusion chamber at 37 °C. Then the common artery was cannulated with a 31-gauge cannula and perfused with a solution containing the muscarinic receptor agonist (CCh, 0.3 μm) plus β-adrenergic agonist (IPR, 1.0 μm). The SMG duct was placed in a calibrated glass capillary tube. The perfusion rate was kept at 1.0 ml/min with a peristaltic pump (IPC-8, ISMATEC, Wertheim, Germany). The ex vivo perfusion solution contained 4.3 mm KCl, 120 mm NaCl, 25 mm NaHCO3, 1 mm CaCl2, 1 mm MgCl2, 5 mm glucose, and 10 mm Hepes, pH 7.2 (gassed with 95% O2, 5% CO2). Secreted saliva samples were collected and stored at −80 °C until analyzed.
Ion concentrations in SMG secretions
Sodium and potassium concentrations were analyzed by atomic absorption spectroscopy (3030 spectrophotometer, PerkinElmer Life Sciences). Chloride activity was measured using a chloride electrode (9617BNWP, Thermo Fisher Scientific) with a 4-Star Plus pH/ISE Benchtop Multiparameter Meter (Thermo Fisher Scientific).
Sample collection and DNA extraction for microbiome analysis
To allow a comprehensive sampling of oral mucosal surfaces, we collected oral mucosal samples from 8-week and 12-week mice. We utilized ultra-fine polystyrene swabs to obtain microbial samples from the murine oral cavity for 30 s, and then these samples were placed in 150 μl of TE buffer and stored at −80 °C until processing. DNA was extracted following a modified protocol of the DNeasy Blood and Tissue Kit (Qiagen) as described previously (41).
16S rRNA gene-based sequencing and bioinformatic analyses
Library generation was performed using Eppendorf liquid-handling robots. The V4 region of the 16S rDNA gene (515F-806R) was sequenced for oral mucosal samples, generating paired-end, overlapping reads on the Illumina MiSeq platform (59). Sequencing data were processed using the software mothur (60). Briefly, reads were quality-filtered and assembled into contigs, and only reads from 200–400 bp in length were kept. Then our processing continued following the MiSeq SOP pipeline as described (61). OTUs were defined at a 97% similarity, and genus-level taxonomical classification was achieved. However, we further informed our classification down to species level through blasting the reference sequence of each OTU against the NCBI 16S rRNA database using BLAST, as described previously (41). β-Diversity was assessed using the θ YC distance. α-Diversity was evaluated using the nonparametric version of the Shannon diversity index. Both diversity measurements were calculated as implemented in mothur. All sequence data from this study have been submitted to the NCBI SRA database under SRA accession number PRJNA547610.
Statistical analysis
Error bars represent S.D. for values obtained from three or more mice. Student's t test and Mann–Whitney test were used to calculate p values. For microbiome analyses, AMOVA was used to test for differences in community structure. LEfSe (62) was used to test for differences in relative abundance of taxa, considering 0.05 as the α value for statistical testing.
Author contributions
G. P. and K. G. T. H. conceptualization; G. P., E. T., and L. A. data curation; G. P., L. A., T. Munemasa, T. Mukaibo, E. T., and K. G. T. H. formal analysis; G. P. validation; G. P., E. T., T. Munemasa, T. Mukaibo, and K. G. T. H. investigation; G. P., L. A., E. T., T. Munemasa, and T. Mukaibo visualization; G. P., E. T., and K. G. T. H. methodology; G. P. and K. G. T. H. writing-original draft; E. T. and K. G. T. H. supervision; K. G. T. H. resources; K. G. T. H. funding acquisition; K. G. T. H. project administration.
Supplementary Material
Acknowledgments
We would thank our colleagues for many helpful discussions. We also thank the NCI Microbiome and Genetics Sequencing Core for 16S sequencing and processing.
This work was supported by Intramural Research Program of the NIDCR, National Institutes of Health, Grants Z01-DE000713 (to K. G. T. H.) and ZIA-DE000738 (to James E. Melvin). This research was also supported in part by the NIDCR Veterinary Resources Core Grant ZIC DE000740-05, the NIDCR Imaging Core Grant DE000750-01, and Chilean National Fund for Scientific and Technologic Development (FONDECYT) Grant 11180505 (to L. A). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3 and Figs. S1–S10.
All sequence data from this study have been submitted to the NCBI SRA database under SRA accession number PRJNA547610.
- Galnt
- UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases
- PNA
- peanut agglutinin
- SMG
- submandibular gland
- MAA
- Maackia amurensis agglutinin
- OTU
- operational taxonomic unit
- HFTC
- hyperphosphatemic familial tumoral calcinosis
- NM
- neuraminidase
- qPCR
- semiquantitative real-time PCR
- LEfSe
- linear discriminant analysis effect size
- E
- embryonic day
- P
- postnatal day
- ER
- endoplasmic reticulum
- PCoA
- principal coordinates analysis
- AMOVA
- analysis of molecular variance
- PFA
- paraformaldehyde
- DIG
- digoxigenin
- TRITC
- tetramethylrhodamine isothiocyanate
- contig
- group of overlapping clones
- θ YC
- θ Yue and Clayton
- TSA
- tyramide signal amplification
- RIPA
- radioimmune precipitation assay.
References
- 1. Kamada N., Chen G. Y., Inohara N., and Núñez G. (2013) Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamont R. J., Koo H., and Hajishengallis G. (2018) The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huttenhower C., Kostic A. D., and Xavier R. J. (2014) Inflammatory bowel disease as a model for translating the microbiome. Immunity 40, 843–854 10.1016/j.immuni.2014.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamont R. J., and Hajishengallis G. (2015) Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abusleme L., Dupuy A. K., Dutzan N., Silva N., Burleson J. A., Strausbaugh L. D., Gamonal J., and Diaz P. I. (2013) The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 10.1038/ismej.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dutzan N., Kajikawa T., Abusleme L., Greenwell-Wild T., Zuazo C. E., Ikeuchi T., Brenchley L., Abe T., Hurabielle C., Martin D., Morell R. J., Freeman A. F., Lazarevic V., Trinchieri G., Diaz P. I., et al. (2018) A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci. Transl. Med. 10, eaat0797 10.1126/scitranslmed.aat0797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griffen A. L., Beall C. J., Campbell J. H., Firestone N. D., Kumar P. S., Yang Z. K., Podar M., and Leys E. J. (2012) Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185 10.1038/ismej.2011.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johansson M. E., and Hansson G. C. (2016) Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16, 639–649 10.1038/nri.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johansson M. E., Sjövall H., and Hansson G. C. (2013) The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10, 352–361 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Culp D. J., Robinson B., Cash M. N., Bhattacharyya I., Stewart C., and Cuadra-Saenz G. (2015) Salivary mucin 19 glycoproteins: innate immune functions in Streptococcus mutans-induced caries in mice and evidence for expression in human saliva. J. Biol. Chem. 290, 2993–3008 10.1074/jbc.M114.597906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frenkel E. S., and Ribbeck K. (2015) Salivary mucins protect surfaces from colonization by cariogenic bacteria. Appl. Environ. Microbiol. 81, 332–338 10.1128/AEM.02573-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frenkel E. S., and Ribbeck K. (2017) Salivary mucins promote the coexistence of competing oral bacterial species. ISME J. 11, 1286–1290 10.1038/ismej.2016.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. An G., Wei B., Xia B., McDaniel J. M., Ju T., Cummings R. D., Braun J., and Xia L. (2007) Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 204, 1417–1429 10.1084/jem.20061929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bergstrom K., Liu X., Zhao Y., Gao N., Wu Q., Song K., Cui Y., Li Y., McDaniel J. M., McGee S., Chen W., Huycke M. M., Houchen C. W., Zenewicz L. A., West C. M., et al. (2016) Defective intestinal mucin-type O-glycosylation causes spontaneous colitis-associated cancer in mice. Gastroenterology 151, 152–164.e11 10.1053/j.gastro.2016.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu J., Wei B., Wen T., Johansson M. E., Liu X., Bradford E., Thomsson K. A., McGee S., Mansour L., Tong M., McDaniel J. M., Sferra T. J., Turner J. R., Chen H., Hansson G. C., et al. (2011) Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J. Clin. Invest. 121, 1657–1666 10.1172/JCI45538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao N., Bergstrom K., Fu J., Xie B., Chen W., and Xia L. (2016) Loss of intestinal O-glycans promotes spontaneous duodenal tumors. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G74–G83 10.1152/ajpgi.00060.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., and Augenlicht L. (2002) Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 10.1126/science.1069094 [DOI] [PubMed] [Google Scholar]
- 18. Wenzel U. A., Magnusson M. K., Rydström A., Jonstrand C., Hengst J., Johansson M. E., Velcich A., Öhman L., Strid H., Sjövall H., Hansson G. C., and Wick M. J. (2014) Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS One 9, e100217 10.1371/journal.pone.0100217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aoki K., Porterfield M., Lee S. S., Dong B., Nguyen K., McGlamry K. H., and Tiemeyer M. (2008) The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J. Biol. Chem. 283, 30385–30400 10.1074/jbc.M804925200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lang T., Hansson G. C., and Samuelsson T. (2007) Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. U.S.A. 104, 16209–16214 10.1073/pnas.0705984104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., Hollingsworth M. A., and Clausen H. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals: one subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 277, 22623–22638 10.1074/jbc.M202684200 [DOI] [PubMed] [Google Scholar]
- 22. Ten Hagen K. G., Tran D. T., Gerken T. A., Stein D. S., and Zhang Z. (2003) Functional characterization and expression analysis of members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family from Drosophila melanogaster. J. Biol. Chem. 278, 35039–35048 10.1074/jbc.M303836200 [DOI] [PubMed] [Google Scholar]
- 23. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 10.1093/glycob/cwr182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tran D. T., and Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 10.1074/jbc.R112.418558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Revoredo L., Wang S., Bennett E. P., Clausen H., Moremen K. W., Jarvis D. L., Ten Hagen K. G., Tabak L. A., and Gerken T. A. (2016) Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 26, 360–376 10.1093/glycob/cwv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian E., and Ten Hagen K. G. (2006) Expression of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology 16, 83–95 10.1093/glycob/cwj051 [DOI] [PubMed] [Google Scholar]
- 27. Ten Hagen K. G., and Tran D. T. (2002) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J. Biol. Chem. 277, 22616–22622 10.1074/jbc.M201807200 [DOI] [PubMed] [Google Scholar]
- 28. Tian E., Hoffman M. P., and Ten Hagen K. G. (2012) O-Glycosylation modulates integrin and FGF signalling by influencing the secretion of basement membrane components. Nat. Commun. 3, 869 10.1038/ncomms1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian E., Stevens S. R., Guan Y., Springer D. A., Anderson S. A., Starost M. F., Patel V., Ten Hagen K. G., and Tabak L. A. (2015) Galnt1 is required for normal heart valve development and cardiac function. PLoS One 10, e0115861 10.1371/journal.pone.0115861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tian E., and Ten Hagen K. G. (2007) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J. Biol. Chem. 282, 606–614 10.1074/jbc.M606268200 [DOI] [PubMed] [Google Scholar]
- 31. Tran D. T., Zhang L., Zhang Y., Tian E., Earl L. A., and Ten Hagen K. G. (2012) Multiple members of the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J. Biol. Chem. 287, 5243–5252 10.1074/jbc.M111.306159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang L., Syed Z. A., van Dijk Härd I., Lim J. M., Wells L., and Ten Hagen K. G. (2014) O-Glycosylation regulates polarized secretion by modulating Tango1 stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7296–7301 10.1073/pnas.1322264111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L., Tran D. T., and Ten Hagen K. G. (2010) An O-glycosyltransferase promotes cell adhesion during development by influencing secretion of an extracellular matrix integrin ligand. J. Biol. Chem. 285, 19491–19501 10.1074/jbc.M109.098145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang L., Turner B., Ribbeck K., and Ten Hagen K. G. (2017) Loss of the mucosal barrier alters the progenitor cell niche via Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling. J. Biol. Chem. 292, 21231–21242 10.1074/jbc.M117.809848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., and Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 10.1038/ng1358 [DOI] [PubMed] [Google Scholar]
- 36. Ichikawa S., Sorenson A. H., Austin A. M., Mackenzie D. S., Fritz T. A., Moh A., Hui S. L., and Econs M. J. (2009) Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology 150, 2543–2550 10.1210/en.2008-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miyazaki T., Mori M., Yoshida C. A., Ito C., Yamatoya K., Moriishi T., Kawai Y., Komori H., Kawane T., Izumi S., Toshimori K., and Komori T. (2013) Galnt3 deficiency disrupts acrosome formation and leads to oligoasthenoteratozoospermia. Histochem. Cell Biol. 139, 339–354 10.1007/s00418-012-1031-3 [DOI] [PubMed] [Google Scholar]
- 38. Zara J., Hagen F. K., Ten Hagen K. G., Van Wuyckhuyse B. C., and Tabak L. A. (1996) Cloning and expression of mouse UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3. Biochem. Biophys. Res. Commun. 228, 38–44 10.1006/bbrc.1996.1613 [DOI] [PubMed] [Google Scholar]
- 39. Gresik E. W. (1975) The postnatal development of the sexually dimorphic duct system and of amylase activity in the submandibular glands of mice. Cell Tissue Res. 157, 411–422 10.1007/bf00225529 [DOI] [PubMed] [Google Scholar]
- 40. Tabak L. A. (1995) In defense of the oral cavity: structure, biosynthesis, and function of salivary mucins. Annu. Rev. Physiol. 57, 547–564 10.1146/annurev.ph.57.030195.002555 [DOI] [PubMed] [Google Scholar]
- 41. Abusleme L., Hong B. Y., Hoare A., Konkel J. E., Diaz P. I., and Moutsopoulos N. M. (2017) Oral microbiome characterization in murine models. Bio. Protoc. 7, e2655 10.21769/BioProtoc.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Y., Gao H., Mihindukulasuriya K. A., La Rosa P. S., Wylie K. M., Vishnivetskaya T., Podar M., Warner B., Tarr P. I., Nelson D. E., Fortenberry J. D., Holland M. J., Burr S. E., Shannon W. D., Sodergren E., and Weinstock G. M. (2013) Biogeography of the ecosystems of the healthy human body. Genome Biol. 14, R1 10.1186/gb-2013-14-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geisler C., and Jarvis D. L. (2011) Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 21, 988–993 10.1093/glycob/cwr080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cross B. W., and Ruhl S. (2018) Glycan recognition at the saliva–oral microbiome interface. Cell Immunol. 333, 19–33 10.1016/j.cellimm.2018.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Denny P. A., and Denny P. C. (1982) Localization of a mouse submandibular sialomucin by indirect immunofluorescence. Histochem. J. 14, 403–408 10.1007/BF01011852 [DOI] [PubMed] [Google Scholar]
- 46. Denny P. A., Denny P. C., and Jenkins K. (1980) Purification and biochemical characterization of a mouse submandibular sialomucin. Carbohydr. Res. 87, 265–274 10.1016/S0008-6215(00)85213-1 [DOI] [PubMed] [Google Scholar]
- 47. Gerken T. A., Jamison O., Perrine C. L., Collette J. C., Moinova H., Ravi L., Markowitz S. D., Shen W., Patel H., and Tabak L. A. (2011) Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 10.1074/jbc.M111.218701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoare A., Marsh P. D., and Diaz P. I. (2017) Ecological therapeutic opportunities for oral diseases. Microbiol. Spectr. 5 10.1128/microbiolspec.BAD-0006-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosier B. T., Marsh P. D., and Mira A. (2018) Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380 10.1177/0022034517742139 [DOI] [PubMed] [Google Scholar]
- 50. Abusleme L., Diaz P. I., Freeman A. F., Greenwell-Wild T., Brenchley L., Desai J. V., Ng W. I., Holland S. M., Lionakis M. S., Segre J. A., Kong H. H., and Moutsopoulos N. M. (2018) Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight 3, 122061 10.1172/jci.insight.122061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moutsopoulos N. M., Chalmers N. I., Barb J. J., Abusleme L., Greenwell-Wild T., Dutzan N., Paster B. J., Munson P. J., Fine D. H., Uzel G., and Holland S. M. (2015) Subgingival microbial communities in leukocyte adhesion deficiency and their relationship with local immunopathology. PLoS Pathog. 11, e1004698 10.1371/journal.ppat.1004698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kiessling L. L., and Grim J. C. (2013) Glycopolymer probes of signal transduction. Chem. Soc. Rev. 42, 4476–4491 10.1039/c3cs60097a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Skoog E. C., Sjöling Å., Navabi N., Holgersson J., Lundin S. B., and Lindén S. K. (2012) Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One 7, e36378 10.1371/journal.pone.0036378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heo S. M., Choi K. S., Kazim L. A., Reddy M. S., Haase E. M., Scannapieco F. A., and Ruhl S. (2013) Host defense proteins derived from human saliva bind to Staphylococcus aureus. Infect. Immun. 81, 1364–1373 10.1128/IAI.00825-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ligtenberg A. J., Walgreen-Weterings E., Veerman E. C., de Soet J. J., de Graaff J., and Amerongen A. V. (1992) Influence of saliva on aggregation and adherence of Streptococcus gordonii HG 222. Infect. Immun. 60, 3878–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moshier A., Reddy M. S., and Scannapieco F. A. (1996) Role of type 1 fimbriae in the adhesion of Escherichia coli to salivary mucin and secretory immunoglobulin A. Curr. Microbiol. 33, 200–208 10.1007/s002849900100 [DOI] [PubMed] [Google Scholar]
- 57. Narimatsu Y., Joshi H. J., Nason R., Van Coillie J., Karlsson R., Sun L., Ye Z., Chen Y. H., Schjoldager K. T., Steentoft C., Furukawa S., Bensing B. A., Sullam P. M., Thompson A. J., Paulson J. C., et al. (2019) An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell 75, 394–407.e5 10.1016/j.molcel.2019.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Romanenko V. G., Nakamoto T., Srivastava A., Begenisich T., and Melvin J. E. (2007) Regulation of membrane potential and fluid secretion by Ca2+-activated K+ channels in mouse submandibular glands. J. Physiol. 581, 801–817 10.1113/jphysiol.2006.127498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., and Knight R. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Lesniewski R. A., Oakley B. B., Parks D. H., Robinson C. J., Sahl J. W., Stres B., Thallinger G. G., Van Horn D. J., and Weber C. F. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., and Schloss P. D. (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., and Huttenhower C. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.